Simulation & design of molecular & soft matter systems
Research
Nanoscopic understanding and control of molecules and materials is pivotal to address urgent challenges in biomedicine and sustainability. With this idea in mind, we develop new computational methods to unravel complex molecular processes occurring at unfathomable length- and timescales: too quick and small for experiments, too slow and large for traditional simulations. We strive to close this gap by boosting molecular dynamics (MD) simulations of larger systems and longer timescales, while collaborating with experimentalists who work in the opposite direction, as well as with other disciplines. Together, we build robust methodologies to answer today’s big scientific and societal challenges.
We build robust methodologies to accelerate simulation and design:
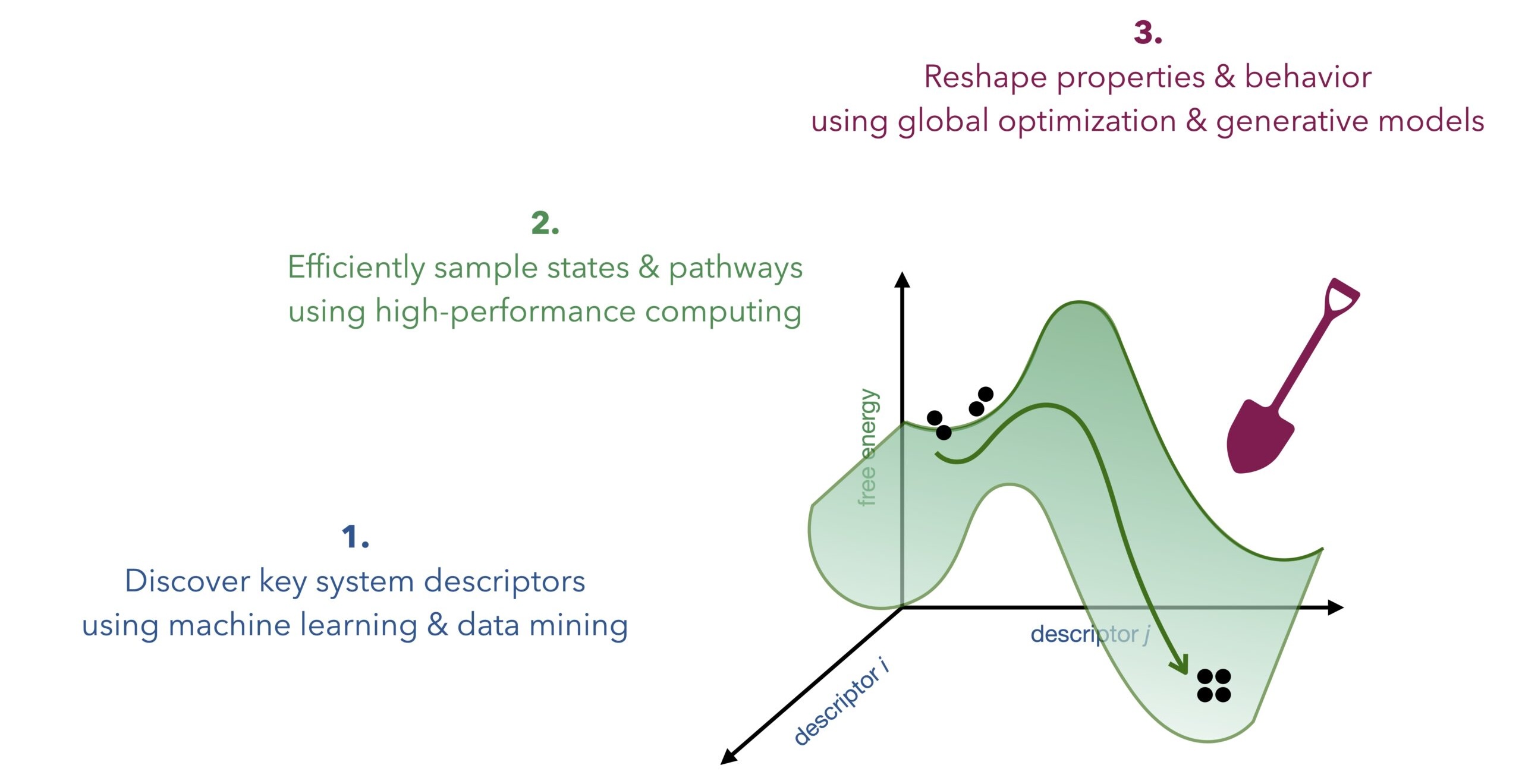
1. via machine learning (ML) and data mining, we find key system descriptors of molecular systems—i.e., order parameters, collective variables or reaction coordinates—giving an interpretable view of their high-dimensional functional dynamics.
2. via descriptor-based enhanced sampling MD simulations and high-performance computing, we efficiently sample (meta)stable states and mechanistic pathways and quantify their free energies, i.e., probabilities.
3. via high-throughput free-energy calculations guided by evolutionary, Bayesian and ML approaches, we reshape free-energy landscapes to design specific behavior and properties, thus predicting and explaining experiments.
Health
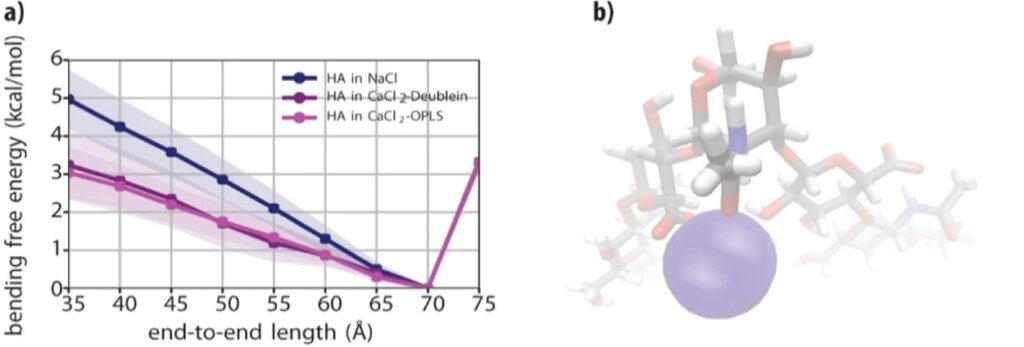
Exploring the ion-responsive mechanical properties of biopolymers
PhD researcher: Eline Kempkes
Erasmus MSc intern: Lisa Roether
Collaborators: G. Giubertoni and S. Woutersen (Molecular Photonics)
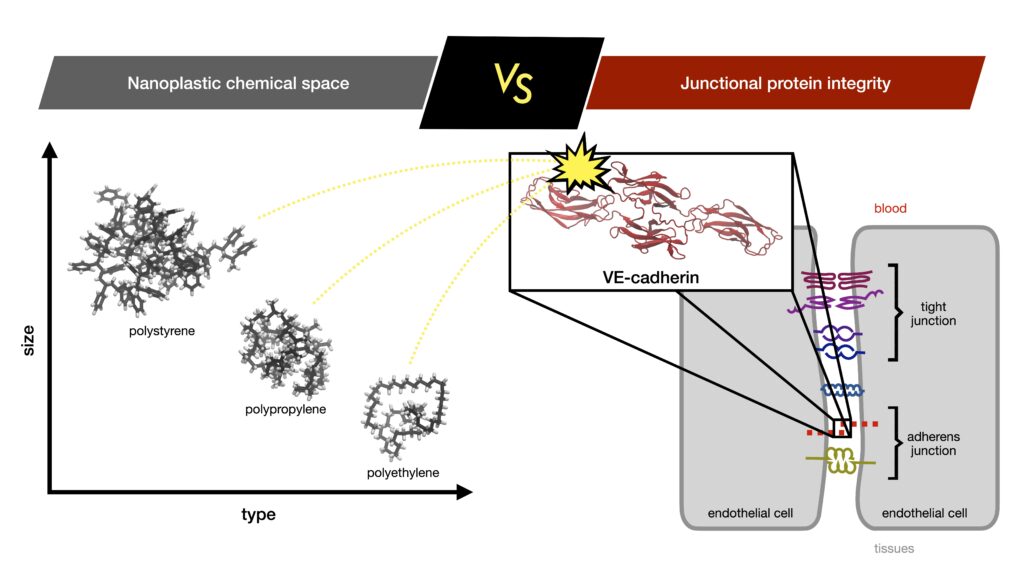
Impact of nanoplastics in protein structure and function
Alumnus MSc Chemistry: Filip Kopczyński
Sustainability
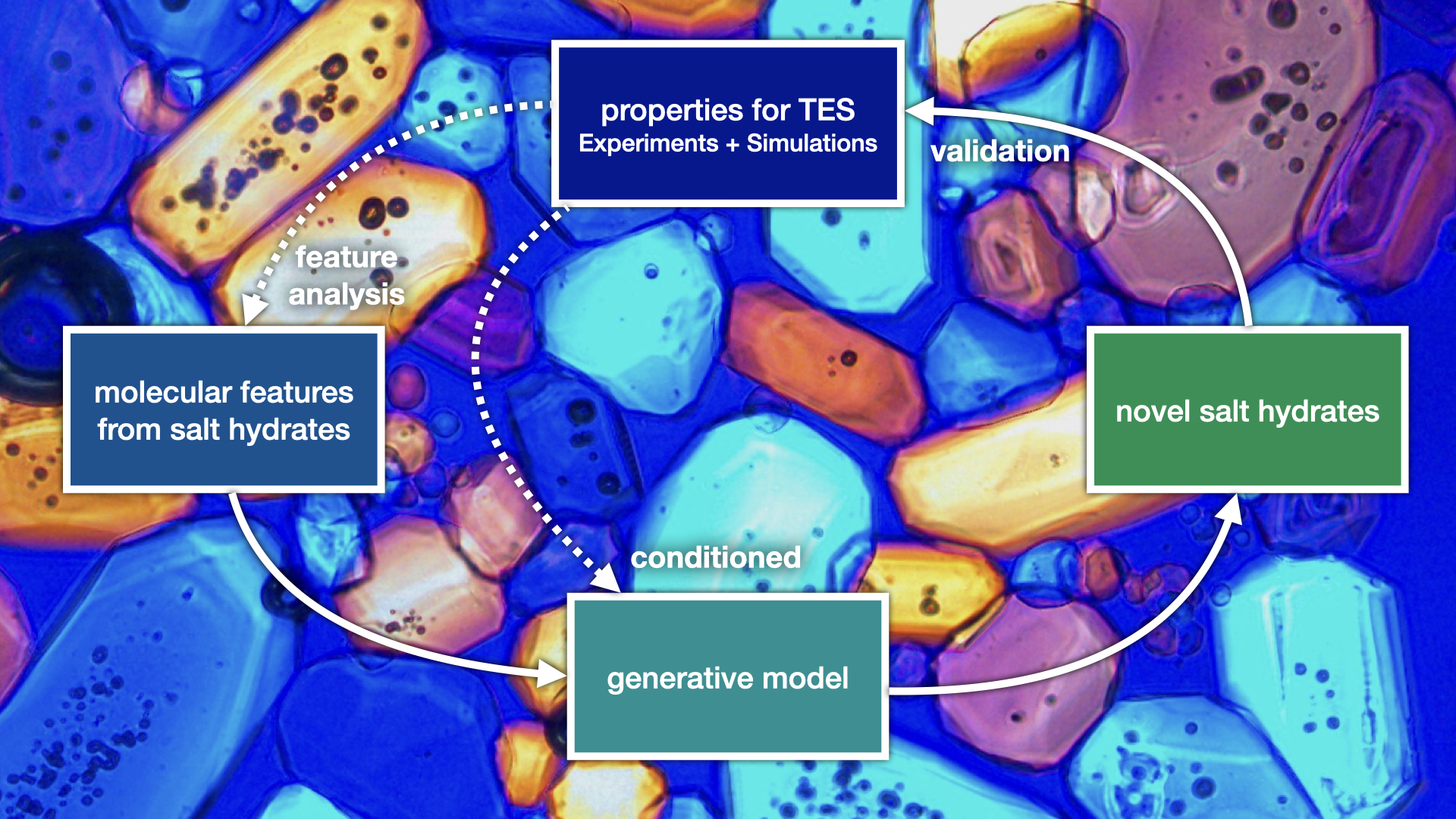
Salt hydrates for thermal energy storage (AI4SMM: Artificial Intelligence for Sustainable Molecules and Materials)
Postdoctoral researcher: Alexander Korotkevich
Co-supervisors: N. Shahidzadeh (Soft Matter) and S. Woutersen (Molecular Photonics)
Machine Learning-based models of plant protein mixtures for sustainable food design (AI4SMM)
Postdoctoral researcher: Maxim Brodmerkel
Co-supervisors: P. Bolhuis (Computational Chemistry), H. van Hoof (Amsterdam ML Lab), S. Jabbari-Farouji (Computational Soft Matter), F. Quattrocchio (Plant Development and EpiGenetics) and P. Shall (Soft Matter)
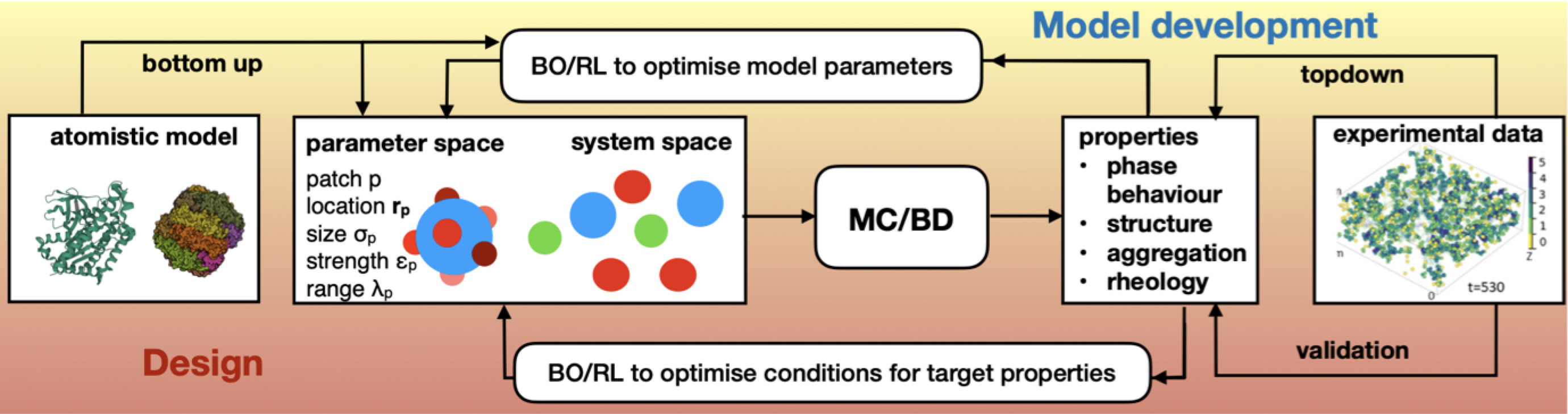
Methodology
Inverse design of self-assembling materials with dynamic design parameters
Alumnus MSc Physics: Nathanyel Schut
Boltzmann generators for molecular design
PhD researcher: Eline Kempkes
Collaborators: T. Bakker, H. van Hoof, M. Welling (Amsterdam ML Lab)
Novel phase behavior
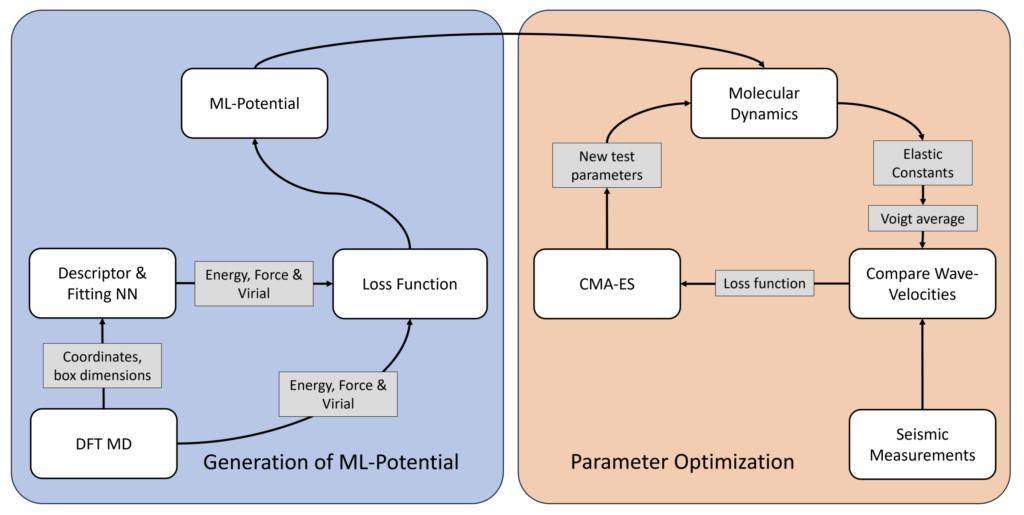
Surrogate modelling of iron at Earth’s inner core extreme conditions by quantum simulations and machine learning
Alumnus MSc Computational Science: Jari Hoffmann
Collaborators: S. Talavera-Soza (Institute of Geophysics and Planetary Physics, UC San Diego).
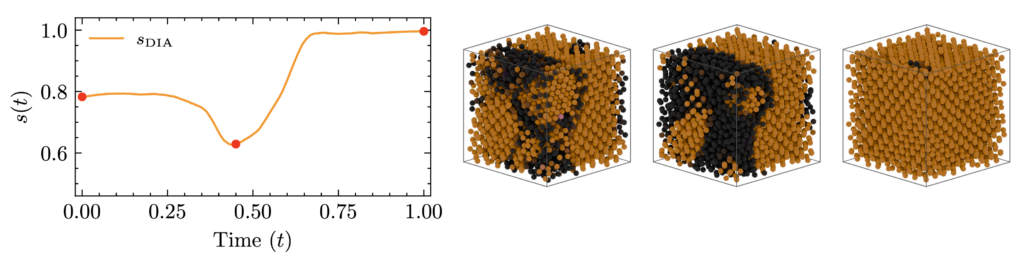
Inverse design of binary quasicrystals
Co-promoted PhD researcher: Edwin Bedolla Montiel
Promotor: Marjolein Dijkstra (Soft Condensed Matter, Utrecht University)
Inverse design of zeolitic phases
Co-supervised PhD researcher: Chaohong Wang
Promotor: Marjolein Dijkstra (Soft Condensed Matter, Utrecht University)
Publications
Vacancies
No vacancies at the moment.
MSc and BSc projects
Interested in designing new molecules and materials via simulation, machine learning and data mining? Contact me.
Bio
I studied Engineering Physics at Tecnológico de Monterrey in Mexico, where I also worked for the cement and steel industry. I later completed a MSc in Computational Science and Engineering at TU München. For my thesis, I worked on adaptive quantum mechanics/molecular mechanics simulations of proton transfers and Bayesian inference of reaction coordinates in the group of Prof. Karsten Reuter. Supported by a personal fellowship, I completed my PhD cum laude in the group of Dr Bernd Ensing in HIMS Computational Chemistry. I focused on developing and applying path-based methods to understand complex chemical and conformational processes in a wide variety of biomolecules. Additionally, I worked on machine learning approaches to discover optimal descriptors for molecular transitions. Subsequently, I joined the Soft Condensed Matter group of Prof. Marjolein Dijkstra at Utrecht University, where I worked on bottom-up design of self-assembly in soft and porous materials at colloidal scales.
Since 2023, I am an assistant professor in the Computational Soft Matter Lab. My current interests include the design of complex free-energy landscapes in molecular and colloidal systems for applications in sustainability and biomedicine. Also in 2023, I became part of the Research Priority Area in Artificial Intelligence for Sustainable Molecules and Materials, co-leading two projects. In 2024, I obtained the prestigious NWO Veni grant to focus on predicting nanoplastic risks to animal junctional proteins. I am an active member of the HIMS Diversity team. Since November 2024, I am a member of the Amsterdam Young Academy.
Links
Artificial Intelligence for Sustainable Molecules and Materials
