Advanced computational methods and machine learning

Dr. Tristan Bereau
Computational Chemistry Group
Computational high-throughput screening in soft matter
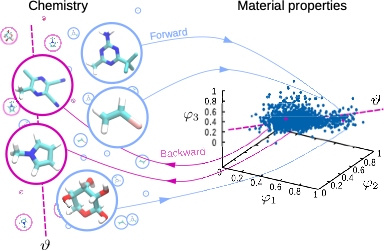
High-throughput screening (see Figure 1) experiments have provided a remarkable body of insight and technological applications in the many fields of materials design—from alloys to drug design. In parallel, recent advances in hardware and algorithms have enabled the development of high-throughput screening from computational methods. In particular, a number of recent applications have demonstrated the use of quantum-chemistry calculations to infer relation-ships regarding electronic properties in hard-condensed matter. Similar computational developments regarding thermodynamic properties had so far remained unexplored. The major technical difficulty was to find a method that would allow a good balance between accuracy and computational cost: obtaining well-converged thermodynamics (such as free energies) with limited computational investment to allow the characterization of many compounds. In a recent work, we have presented a novel methodology that relies on coarse-grained simulations to enable such high-throughput free-energy calculations. The large speed-up compared to atomistic simulations is the effect of both a more efficient sampling of conformational space (i.e., the free-energy calculation) as well as a reduction of chemical space (i.e., the number of possible molecules). The latter effect means that many molecules map to a single coarse-grained representation. As such, this loss of chemical detail of the coarser model translates into a significant advantage: it reduces the number of molecules that ought to be considered when screening across chemistry. This makes for an elegant and systematic strategy to explore the diversity of chemical space.
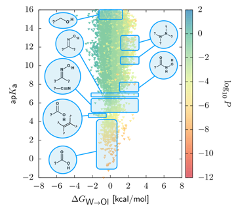
We have considered the permeation of small molecules across a phospholipid bilayer, and focused on predicting the coefficient of passive permeation. Despite great interest, extracting the passive permeation has remained challenging both experimentally and from computer simulations. The coarse-grained simulations have allowed us to describe the permeability coefficients for hundreds of thousands of compounds. We have generated a smooth permeability surface as a function of two parameters: the partitioning free energy and the ionization constant (pKa). This effectively provides a direct structure-property relationship for this phenomenon (Figure 2). Abundant validation both in and beyond the molecular weight considered here makes clear of the robustness of our methodology.
Physically-motivated machine learning for multiscale molecular simulations
Advanced statistical methods are rapidly impregnating many scientific fields, offering new perspectives on long-standing problems. In materials science, data-driven methods are already bearing fruit in various disciplines, such as hard condensed matter or inorganic chemistry, as well as soft matter to a smaller extent.
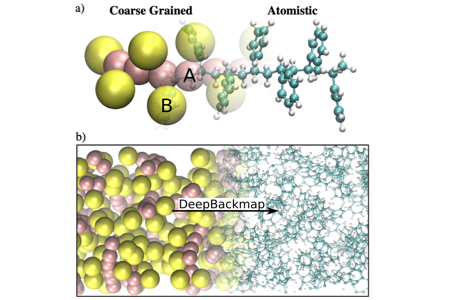
When coupling machine learning to molecular simulations, many problems of interest display dauntingly-large interpolation spaces, limiting their immediate application without undesired artifacts (e.g., extrapolation). The incorporation of physical information, such as conserved quantities, symmetries, and constraints, can play a decisive role in reducing the interpolation space. Conversely, physics can help determine whether a machine-learning prediction should be trusted, acting as a more robust alternative to the predictive variance.
We show how incorporating physics in machine-learning models can help connect resolutions in multiscale modeling. Applications include force-field parametrization, automated dimensionality reduction and clustering, and generative models to reintroduce atomistic detail in coarse-grained configurations.
Advanced Monte Carlo methods

Dr. David Dubbeldam
Computational Chemistry Group
In statistical mechanics, open systems are defined as a finite volume V with walls permeable to molecules in equilibrium with an infinite reservoir of the same temperature and activity. Examples are the grand-canonical ensemble, the Gibbs ensembles, and reaction ensembles. In open systems, the chemical potential is fixed and hence the conjugate variable, the number of particles, fluctuates. Methods that rely on the insertion and deletion of molecules and on the spontenous formation of empty insertion cavities start to fail at high density and/or low temperature. Recent insertion/deletion methodologies do not rely on sponteneous formation of insertion cavities and are hence applicable to low temperature and high densities. Also, sometimes insertion/deletion can be avoided altogether by sampling a finite spatial region from a larger system. We work on simulation methods that efficiently sample open systems.
Trajectory based methods for rare event sampling

Prof. dr. Peter Bolhuis
Computational Chemistry Group
When confronted with a rare event one often resorts to biased sampling. An alternative is to use transition path sampling (TPS) which efficiently samples trajectories between two predefined states without prior knowledge of the reaction coordinates. Rates can be computed with Trantion interface sampling. Knowledge of the reaction or transition mechanism is often an important goal for molecular simulations. Using TPS there are several ways to extract reaction coordanates using variants of Machine Learning. We are actively developing such path based methodology .