Biography
Ioana studied Engineering Physics (graduated summa cum laude) as well as Business Administration at the Babes-Bolyai University, Romania. In 2011 she obtained her MSc in Computational Physics and her MBA. In December 2015 she was awarded her PhD in Computational Biophysics from the University of Twente in the Netherlands, where she developed coarse grained models for intrinsically disordered proteins. As a postdoctoral researcher she switched focus towards higher resolution models. During her stay at the University of Darmstadt, Germany, she investigated the molecular mechanisms underlying fibril elongation (2016/2017). She then became a Peter und Traudl Engelhorn Postdoctoral Research Fellow at the University of Zürich, Switzerland. Since February 2022 she is an Assistant Professor in the Computational Chemistry group.
Research interests
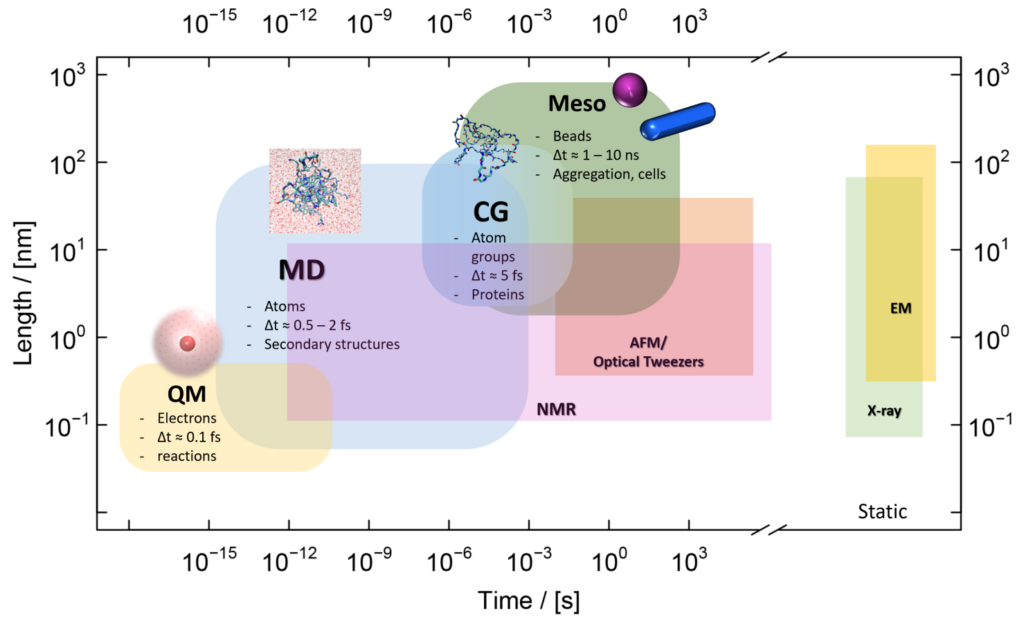
Multiscale simulations of biomolecular systems
We use multiscale simulations and interdisciplinary approaches to investigate the properties of self-assembled macromolecules. We focus on the thermodynamics and kinetics of protein folding and aggregation and rational design of agents to modulate the functional properties of proteins and nanoparticles. We atomistic and coarse-grained simulations combined with methodologies from biophysics and biochemistry.
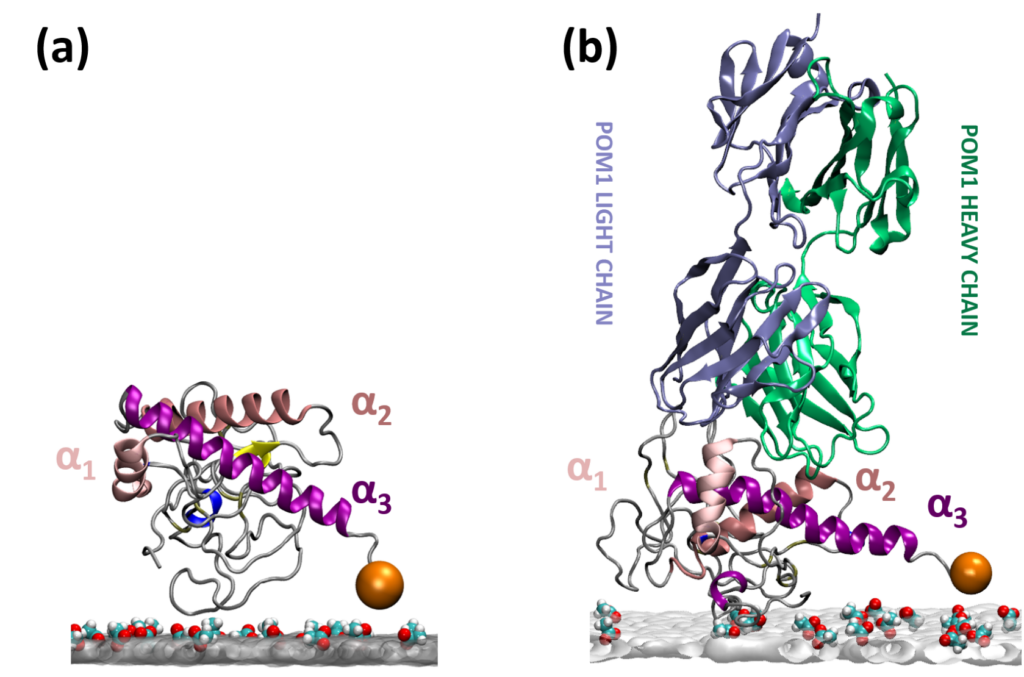
Peptide based therapeutics for neurodegenerative diseases
We use rational design and computer simulations combined with experimental validation to develop a strategy to block the toxicity triggered by proteins associated with neurodegenerative diseases (e.g. prion diseases). In particular, we design peptides to bind to the healthy prion protein with high specificity and probe their efficacy by enhanced sampling molecular dynamics simulations.
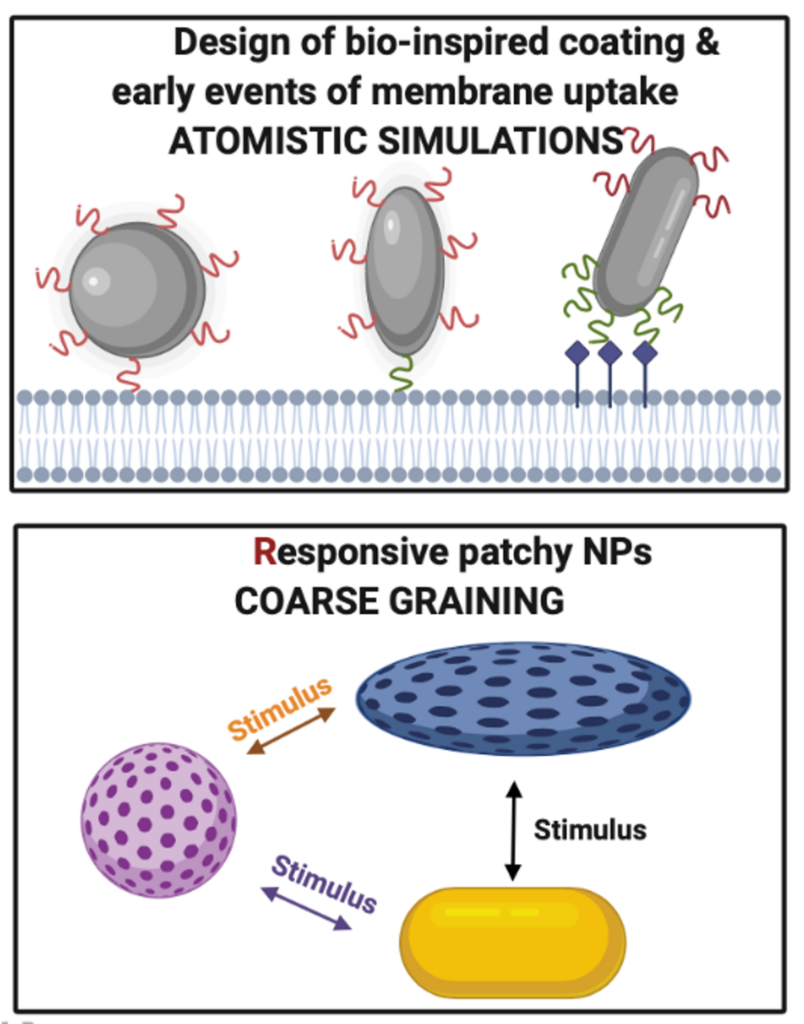
Nanocarriers and non-invasive therapeutics
Nanoparticles (NPs) are used to efficiently deliver drugs into diseased cells. A rate limiting step in the successful transport of NPs into the target cell is the interaction with the membrane.
We design bio-inspired ligands for nanoparticles and explore the early events at the membrane interface via atomistic simulations.
To capture the full cellular uptake mechanisms, we design coarse-grained models able to capture the intrinsic flexibility of the nanoparticles as well as the targeting specificity introduced by the ligands.
