New paper by Tamika van ‘t Hoff
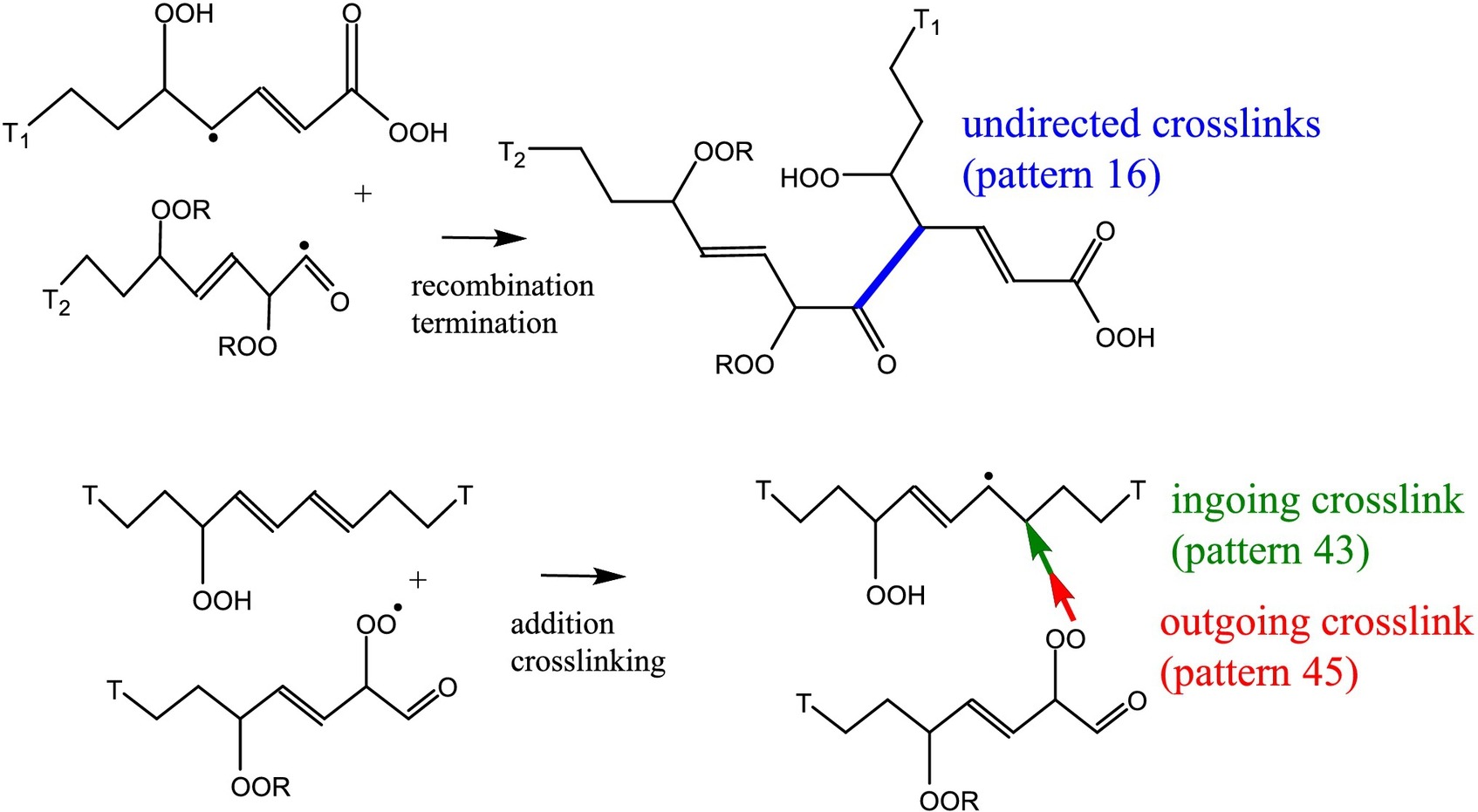
Tamika has published a new paper about the “Experimental validation of a reaction network model for autoxidation of linoleate esters“. CompChem alumni Yuliia Orlova and Becca Harmon are also authors, as well as HIMS researchers Joen Hermans , Alessa Gambardella, and Piet Iedema. Congrats, Tamika!
Read more