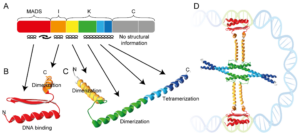
Protein folding and aggregation are fundamental biological processes that up to date remain elusive. The complex nature of the living organism relies on interactions between peptides to fulfill their natural function. The aim of this project is to investigate the structural and dynamic properties of functional amyloidogenic peptides by means of molecular dynamics simulations. In particular, we will use enhanced sampling techniques [1] to explore conformational free energy landscape of the monomers in the Staphylococcus aureus PSMs peptide family.
[1] I.M. ILIE et al. Intrinsic conformational preferences and interactions in α-synuclein fibrils: insights from molecular dynamics simulations J.C.T.C., 14, 3298-3310 (2018)