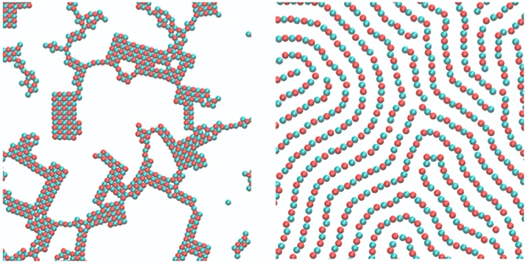
Continuous injection of energy into many-body systems can lead to formation of novel non-equilibrium structures which are inaccessible in equilibrium assembly. From the statistical physics perspective, the nature of dynamical order-disorder transitions in non-equilibrium structure formation is poorly understood. In this project, we would like to gain fundamental insights into the process of dynamic assembly by performing Brownian dynamic simulations of simple colloidal model systems driven out of equilibrium through oscillatory interaction potentials or consumption [1] of fuel via dissipative reaction networks [2]. Computer simulations allow us to explore the non-equilibrium pathways of structure formation by directly analyzing the ensembles of non-equilibrium trajectories. Elucidating the nature of dynamic self-assembly in model colloidal systems provides us with a scaffold for understanding the more complex dynamic self-assembly processes occurring in a variety of living systems.
References:
[1] Dissipative self-assembly of particles interacting through time-oscillatory potential, M. Tagliazucchi, Emily A. Weiss, and Igal Szleifer, PNAS 111 (27) 9751-9756 (2014).
[2] Out-of-Equilibrium Colloidal Assembly Driven by Chemical Reaction Networks B. G. P. van Ravensteijn, et. al., Langmuir2020, 36, 10639−10656