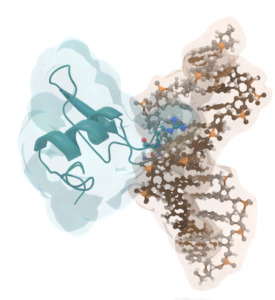
Histones are considered to be the central organizers of eukaryotic chromatin. They are able to wrap DNA around an octameric histone core forming nucleosomes. Many archaea express homologues of these histones, of which some are considered to be the evolutionary predecessors of eukaryotic histones. Recent work of the Molecular and Cellular Biochemistry group at Leiden University showed that the archaeal HMfA and HMfB histones from Methanothermus fervidus can wrap DNA around an ‘endless’ histone protein core to form a hypernucleosome, see Figure 1.
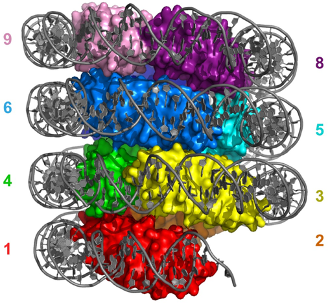
The histone proteins HTkA and HTkB of Thermococcus kodakarensis showed similar behavior, suggesting that the formation of hypernucleosomes is conserved between these histones. Mutagenesis studies showed that the hypernucleosome structure is stabilized by electrostatic stacking interactions, mediated by lysines. A few lysines involved have been found to be prone to acetylation, suggesting a role for post-translational modifications in modulating chromatin structure and therewith gene regulation. Mutating these electrostatic stacking interactions was shown to affect the DNA compacting ability of HTk/HMf histones, DNA binding affinity, and cooperativity of the histone proteins. Remarkably, mutating the stacking lysines in HTkA and HTkB still result in hypernucleosomes, but with a different degree of DNA compaction. This difference, despite the high similarity between HTkA and HTkB suggests that the stacking mutation affects the histone-DNA interaction, possibly mediated by structural changes in the protein close to the DNA interaction surface.
To further explore the nature of the stacking interactions, high resolution in both space and time is required, which can be provided by molecular simulation. In this project you will investigate the stacking interactions in HTkA and HTkB using molecular dynamics simulations. Comparing long (~us) molecular dynamics simulations of the wild type and mutations will reveal whether removing the electrostatic interactions will have structural effects on the histone proteins that go beyond the mere perturbation of stacking interactions. The results of this project can aid in explaining the experimental results in more detail.
This project is in collaboration with Ilias Zarguit and Remus Dame, Molecular and Cellular Biochemistry at Leiden University.
Further reading
Henneman, B. and R. T. Dame (2015). “Archaeal histones: dynamic and versatile genome architects.” Aims Microbiology 1(1): 72-81.
Henneman, B., et al. (2018). “Structure and function of archaeal histones.” PLoS Genet 14(9): e1007582.
Mattiroli, F., et al. (2017). “Structure of histone-based chromatin in Archaea.” Science 357(6351): 609-612.
Henneman, B., et al. (2021). “Mechanical and structural properties of archaeal hypernucleosomes.” Nucleic Acids Res 49(8): 4338-4349.
Alpha-Bazin, B., et al. (2021). “Lysine-specific acetylated proteome from the archaeon Thermococcus gammatolerans reveals the presence of acetylated histones.” J Proteomics 232: 104044